Drug Name Inventor¶
Mitch Fawcett
October 3, 2018
It seems the pharmaceutical industry has devoured all the likely names for new drugs so it is getting harder and harder to come up with new ones.
This Python program attempts to solve that problem using deep learning. A character based language model will be trained using a list of most of the existing drugs to generate new names suitable for marketing.
It assumes another Python program called LoadDrugNames has previously compiled the list of existing drug names to use for training the model. The drug list was found on the NIH Web site and contains the names of about 3,000 drugs. The data engineering that went into extracting the names from the Web site will be skipped over here.

|
We will begin by loading the libraries we'll need plus some functions that are provided in rnn_utils. Specifically; functions such as rnn_forward and rnn_backward.
import numpy as np
from utils import *
import random
data = open('drugnames.csv', 'r').read()
data= data.lower()
chars = list(set(data))
data_size, vocab_size = len(data), len(chars)
drug_count = len(open('drugnames.csv').readlines())
print('There are %d drug names in the training data.' % (drug_count))
print('There are %d total characters and %d unique characters in the training data.' % (data_size, vocab_size))
There are 3047 drug names in the training data. There are 29291 total characters and 27 unique characters in the training data.
The characters are a-z (26 characters) plus the "\n" (or newline character), which indicates the end of the drug name. In the cell below, we create a python dictionary (i.e., a hash table) to map each character to an index from 0-26. We also create a second python dictionary that maps each index back to the corresponding character character. This will help you figure out what index corresponds to what character in the probability distribution output of the softmax layer. Below, char_to_ix and ix_to_char are the python dictionaries.
char_to_ix = { ch:i for i,ch in enumerate(sorted(chars)) }
ix_to_char = { i:ch for i,ch in enumerate(sorted(chars)) }
print(ix_to_char)
print(char_to_ix)
{0: '\n', 1: 'a', 2: 'b', 3: 'c', 4: 'd', 5: 'e', 6: 'f', 7: 'g', 8: 'h', 9: 'i', 10: 'j', 11: 'k', 12: 'l', 13: 'm', 14: 'n', 15: 'o', 16: 'p', 17: 'q', 18: 'r', 19: 's', 20: 't', 21: 'u', 22: 'v', 23: 'w', 24: 'x', 25: 'y', 26: 'z'}
{'\n': 0, 'a': 1, 'b': 2, 'c': 3, 'd': 4, 'e': 5, 'f': 6, 'g': 7, 'h': 8, 'i': 9, 'j': 10, 'k': 11, 'l': 12, 'm': 13, 'n': 14, 'o': 15, 'p': 16, 'q': 17, 'r': 18, 's': 19, 't': 20, 'u': 21, 'v': 22, 'w': 23, 'x': 24, 'y': 25, 'z': 26}
1.2 - Overview of the model¶
Your model will have the following structure:
- Initialize parameters
- Run the optimization loop
- Forward propagation to compute the loss function
- Backward propagation to compute the gradients with respect to the loss function
- Clip the gradients to avoid exploding gradients
- Using the gradients, update your parameter with the gradient descent update rule.
- Return the learned parameters
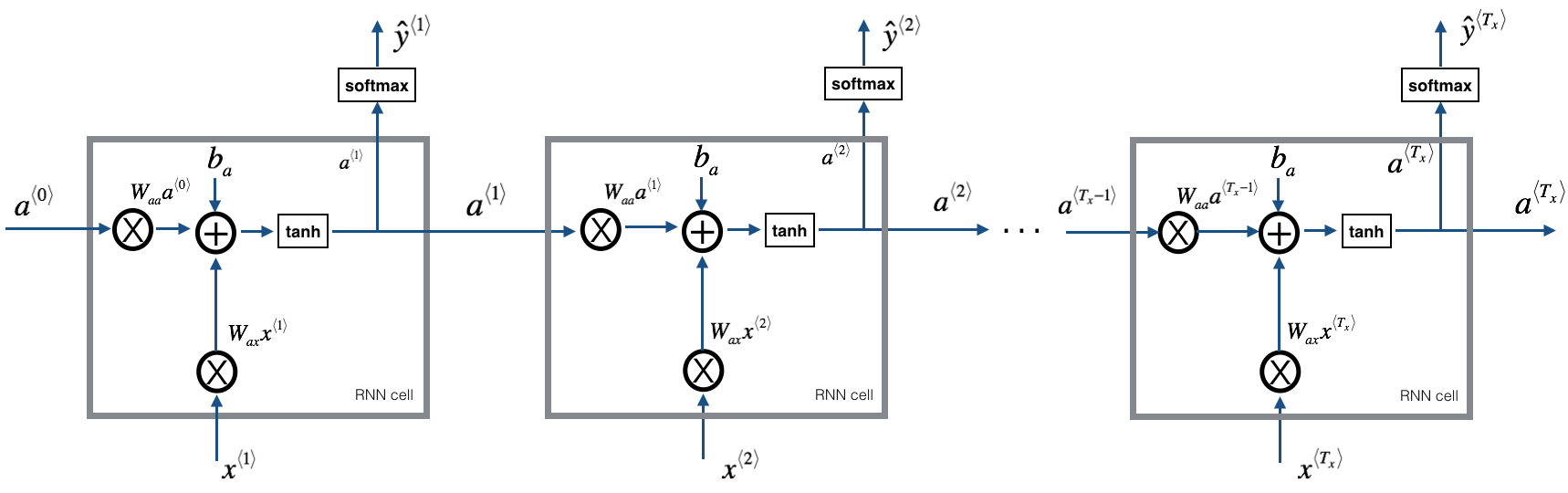
At each time-step, the RNN tries to predict what is the next character given the previous characters. The dataset $X = (x^{\langle 1 \rangle}, x^{\langle 2 \rangle}, ..., x^{\langle T_x \rangle})$ is a list of characters in the training set, while $Y = (y^{\langle 1 \rangle}, y^{\langle 2 \rangle}, ..., y^{\langle T_x \rangle})$ is such that at every time-step $t$, we have $y^{\langle t \rangle} = x^{\langle t+1 \rangle}$.
2 - Building blocks of the model¶
In this part, you will build two important blocks of the overall model:
- Gradient clipping: to avoid exploding gradients
- Sampling: a technique used to generate characters
You will then apply these two functions to build the model.
2.1 - Clipping the gradients in the optimization loop¶
In this section you will implement the clip function that you will call inside of your optimization loop. Recall that your overall loop structure usually consists of a forward pass, a cost computation, a backward pass, and a parameter update. Before updating the parameters, you will perform gradient clipping when needed to make sure that your gradients are not "exploding," meaning taking on overly large values.
In the exercise below, you will implement a function clip that takes in a dictionary of gradients and returns a clipped version of gradients if needed. There are different ways to clip gradients; we will use a simple element-wise clipping procedure, in which every element of the gradient vector is clipped to lie between some range [-N, N]. More generally, you will provide a maxValue (say 10). In this example, if any component of the gradient vector is greater than 10, it would be set to 10; and if any component of the gradient vector is less than -10, it would be set to -10. If it is between -10 and 10, it is left alone.
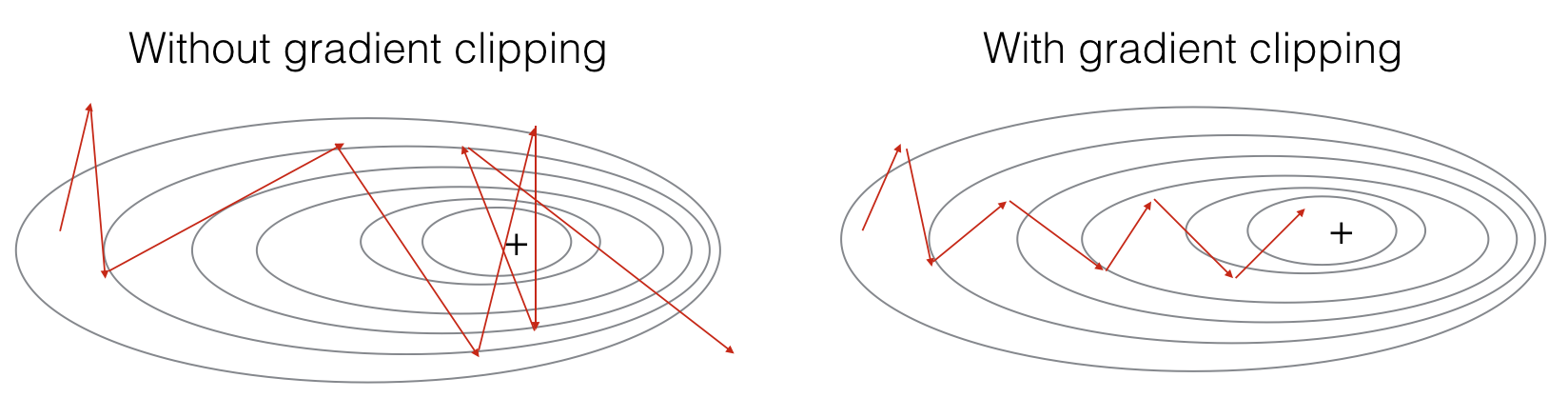
Exercise: Implement the function below to return the clipped gradients of your dictionary gradients. Your function takes in a maximum threshold and returns the clipped versions of your gradients. You can check out this hint for examples of how to clip in numpy. You will need to use the argument out = ....
### GRADED FUNCTION: clip
def clip(gradients, maxValue):
'''
Clips the gradients' values between minimum and maximum.
Arguments:
gradients -- a dictionary containing the gradients "dWaa", "dWax", "dWya", "db", "dby"
maxValue -- everything above this number is set to this number, and everything less than -maxValue is set to -maxValue
Returns:
gradients -- a dictionary with the clipped gradients.
'''
dWaa, dWax, dWya, db, dby = gradients['dWaa'], gradients['dWax'], gradients['dWya'], gradients['db'], gradients['dby']
### START CODE HERE ###
# clip to mitigate exploding gradients, loop over [dWax, dWaa, dWya, db, dby]. (≈2 lines)
for gradient in [dWax, dWaa, dWya, db, dby]:
np.clip(gradient, -1 * maxValue, maxValue, out = gradient)
### END CODE HERE ###
gradients = {"dWaa": dWaa, "dWax": dWax, "dWya": dWya, "db": db, "dby": dby}
return gradients
np.random.seed(3)
dWax = np.random.randn(5,3)*10
dWaa = np.random.randn(5,5)*10
dWya = np.random.randn(2,5)*10
db = np.random.randn(5,1)*10
dby = np.random.randn(2,1)*10
gradients = {"dWax": dWax, "dWaa": dWaa, "dWya": dWya, "db": db, "dby": dby}
gradients = clip(gradients, 10)
print("gradients[\"dWaa\"][1][2] =", gradients["dWaa"][1][2])
print("gradients[\"dWax\"][3][1] =", gradients["dWax"][3][1])
print("gradients[\"dWya\"][1][2] =", gradients["dWya"][1][2])
print("gradients[\"db\"][4] =", gradients["db"][4])
print("gradients[\"dby\"][1] =", gradients["dby"][1])
gradients["dWaa"][1][2] = 10.0 gradients["dWax"][3][1] = -10.0 gradients["dWya"][1][2] = 0.2971381536101662 gradients["db"][4] = [10.] gradients["dby"][1] = [8.45833407]
** Expected output:**
| **gradients["dWaa"][1][2] ** | 10.0 |
| **gradients["dWax"][3][1]** | -10.0 |
| **gradients["dWya"][1][2]** | 0.29713815361 |
| **gradients["db"][4]** | [ 10.] |
| **gradients["dby"][1]** | [ 8.45833407] |
2.2 - Sampling¶
Now assume that your model is trained. You would like to generate new text (characters). The process of generation is explained in the picture below:
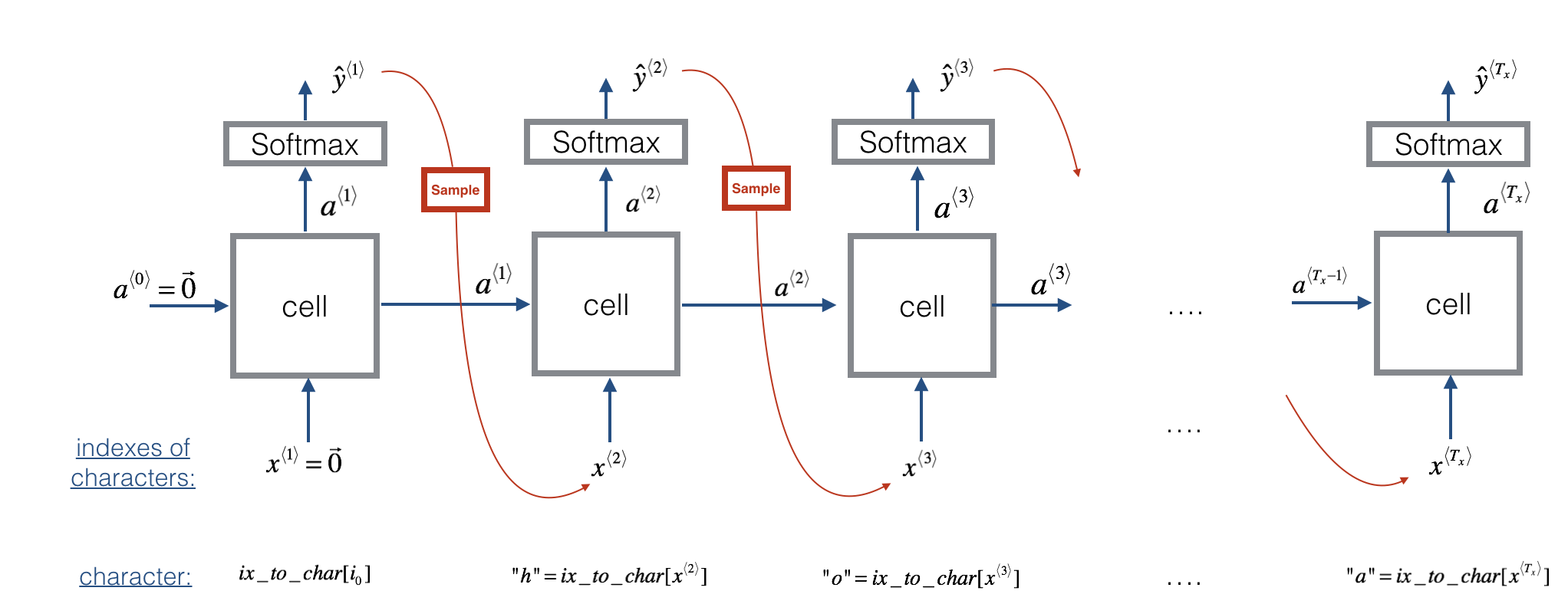
Exercise: Implement the sample function below to sample characters. You need to carry out 4 steps:
Step 1: Pass the network the first "dummy" input $x^{\langle 1 \rangle} = \vec{0}$ (the vector of zeros). This is the default input before we've generated any characters. We also set $a^{\langle 0 \rangle} = \vec{0}$
Step 2: Run one step of forward propagation to get $a^{\langle 1 \rangle}$ and $\hat{y}^{\langle 1 \rangle}$. Here are the equations:
Note that $\hat{y}^{\langle t+1 \rangle }$ is a (softmax) probability vector (its entries are between 0 and 1 and sum to 1). $\hat{y}^{\langle t+1 \rangle}_i$ represents the probability that the character indexed by "i" is the next character. We have provided a softmax() function that you can use.
- Step 3: Carry out sampling: Pick the next character's index according to the probability distribution specified by $\hat{y}^{\langle t+1 \rangle }$. This means that if $\hat{y}^{\langle t+1 \rangle }_i = 0.16$, you will pick the index "i" with 16% probability. To implement it, you can use
np.random.choice.
Here is an example of how to use np.random.choice():
np.random.seed(0)
p = np.array([0.1, 0.0, 0.7, 0.2])
index = np.random.choice([0, 1, 2, 3], p = p.ravel())
This means that you will pick the index according to the distribution:
$P(index = 0) = 0.1, P(index = 1) = 0.0, P(index = 2) = 0.7, P(index = 3) = 0.2$.
- Step 4: The last step to implement in
sample()is to overwrite the variablex, which currently stores $x^{\langle t \rangle }$, with the value of $x^{\langle t + 1 \rangle }$. You will represent $x^{\langle t + 1 \rangle }$ by creating a one-hot vector corresponding to the character you've chosen as your prediction. You will then forward propagate $x^{\langle t + 1 \rangle }$ in Step 1 and keep repeating the process until you get a "\n" character, indicating you've reached the end of the dinosaur name.
# GRADED FUNCTION: sample
def sample(parameters, char_to_ix, seed, max_length):
"""
Sample a sequence of characters according to a sequence of probability distributions output of the RNN
Arguments:
parameters -- python dictionary containing the parameters Waa, Wax, Wya, by, and b.
char_to_ix -- python dictionary mapping each character to an index.
seed -- used for grading purposes. Do not worry about it.
max_length - the maximum number of characters to use in a name
Returns:
indices -- a list of length n containing the indices of the sampled characters.
"""
# Retrieve parameters and relevant shapes from "parameters" dictionary
Waa, Wax, Wya, by, b = parameters['Waa'], parameters['Wax'], parameters['Wya'], parameters['by'], parameters['b']
vocab_size = by.shape[0]
n_a = Waa.shape[1]
### START CODE HERE ###
# Step 1: Create the one-hot vector x for the first character (initializing the sequence generation). (≈1 line)
x = np.zeros(shape = (vocab_size, 1))
# Step 1': Initialize a_prev as zeros (≈1 line)
a_prev = np.zeros(shape = (n_a, 1))
# Create an empty list of indices, this is the list which will contain the list of indices of the characters to generate (≈1 line)
indices = []
# Idx is a flag to detect a newline character, we initialize it to -1
idx = -1
# Loop over time-steps t. At each time-step, sample a character from a probability distribution and append
# its index to "indices". We'll stop if we reach max_length characters (which should be very unlikely with a well
# trained model), which helps debugging and prevents entering an infinite loop.
counter = 0
newline_character = char_to_ix['\n']
while (idx != newline_character and counter != max_length):
# Step 2: Forward propagate x using the equations (1), (2) and (3)
a = np.tanh(np.add(np.matmul(Wax, x), np.matmul(Waa, a_prev)) + b)
z = np.matmul(Wya, a) + by
y = softmax(z)
# print(y)
# print(np.sum(y))
# for grading purposes
np.random.seed(counter+seed)
# Step 3: Sample the index of a character within the vocabulary from the probability distribution y
idx = np.random.choice(vocab_size, p = y.ravel())
### Note - despite the "random.choice" in the name of the above function, it is not randomly choosing
### a letter index. It is choosing the letter index with the highest probability. It will choose the
### index of the element in y that has the highest value.
## print(y[idx])
## print(idx)
## print(ix_to_char[idx])
# Append the index to "indices"
indices.append(idx)
# Step 4: Overwrite the input character as the one corresponding to the sampled index
x = np.zeros(shape = (vocab_size, 1))
x[idx] = 1
# Update "a_prev" to be "a"
a_prev = a
# for grading purposes
seed += 1
counter +=1
### END CODE HERE ###
if (counter == max_length):
indices.append(char_to_ix['\n'])
return indices
np.random.seed(2)
_, n_a = 20, 100
Wax, Waa, Wya = np.random.randn(n_a, vocab_size), np.random.randn(n_a, n_a), np.random.randn(vocab_size, n_a)
b, by = np.random.randn(n_a, 1), np.random.randn(vocab_size, 1)
parameters = {"Wax": Wax, "Waa": Waa, "Wya": Wya, "b": b, "by": by}
indices = sample(parameters, char_to_ix, 0, 20)
print("Sampling:")
print("list of sampled indices:", indices)
print("list of sampled characters:", [ix_to_char[i] for i in indices])
Sampling: list of sampled indices: [12, 17, 24, 14, 13, 9, 10, 22, 24, 6, 13, 11, 12, 6, 21, 15, 21, 14, 3, 2, 0] list of sampled characters: ['l', 'q', 'x', 'n', 'm', 'i', 'j', 'v', 'x', 'f', 'm', 'k', 'l', 'f', 'u', 'o', 'u', 'n', 'c', 'b', '\n']
** Expected output:**
| **list of sampled indices:** |
[12, 17, 24, 14, 13, 9, 10, 22, 24, 6, 13, 11, 12, 6, 21, 15, 21, 14, 3, 2, 1, 21, 18, 24, 7, 25, 6, 25, 18, 10, 16, 2, 3, 8, 15, 12, 11, 7, 1, 12, 10, 2, 7, 7, 11, 5, 6, 12, 25, 0, 0] |
| **list of sampled characters:** |
['l', 'q', 'x', 'n', 'm', 'i', 'j', 'v', 'x', 'f', 'm', 'k', 'l', 'f', 'u', 'o', 'u', 'n', 'c', 'b', 'a', 'u', 'r', 'x', 'g', 'y', 'f', 'y', 'r', 'j', 'p', 'b', 'c', 'h', 'o', 'l', 'k', 'g', 'a', 'l', 'j', 'b', 'g', 'g', 'k', 'e', 'f', 'l', 'y', '\n', '\n'] |
3 - Building the language model¶
It is time to build the character-level language model for text generation.
3.1 - Gradient descent¶
In this section you will implement a function performing one step of stochastic gradient descent (with clipped gradients). You will go through the training examples one at a time, so the optimization algorithm will be stochastic gradient descent. As a reminder, here are the steps of a common optimization loop for an RNN:
- Forward propagate through the RNN to compute the loss
- Backward propagate through time to compute the gradients of the loss with respect to the parameters
- Clip the gradients if necessary
- Update your parameters using gradient descent
Exercise: Implement this optimization process (one step of stochastic gradient descent).
We provide you with the following functions:
def rnn_forward(X, Y, a_prev, parameters):
""" Performs the forward propagation through the RNN and computes the cross-entropy loss.
It returns the loss' value as well as a "cache" storing values to be used in the backpropagation."""
....
return loss, cache
def rnn_backward(X, Y, parameters, cache):
""" Performs the backward propagation through time to compute the gradients of the loss with respect
to the parameters. It returns also all the hidden states."""
...
return gradients, a
def update_parameters(parameters, gradients, learning_rate):
""" Updates parameters using the Gradient Descent Update Rule."""
...
return parameters
# GRADED FUNCTION: optimize
def optimize(X, Y, a_prev, parameters, learning_rate = 0.01):
"""
Execute one step of the optimization to train the model.
Arguments:
X -- list of integers, where each integer is a number that maps to a character in the vocabulary.
Y -- list of integers, exactly the same as X but shifted one index to the left.
a_prev -- previous hidden state.
parameters -- python dictionary containing:
Wax -- Weight matrix multiplying the input, numpy array of shape (n_a, n_x)
Waa -- Weight matrix multiplying the hidden state, numpy array of shape (n_a, n_a)
Wya -- Weight matrix relating the hidden-state to the output, numpy array of shape (n_y, n_a)
b -- Bias, numpy array of shape (n_a, 1)
by -- Bias relating the hidden-state to the output, numpy array of shape (n_y, 1)
learning_rate -- learning rate for the model.
Returns:
loss -- value of the loss function (cross-entropy)
gradients -- python dictionary containing:
dWax -- Gradients of input-to-hidden weights, of shape (n_a, n_x)
dWaa -- Gradients of hidden-to-hidden weights, of shape (n_a, n_a)
dWya -- Gradients of hidden-to-output weights, of shape (n_y, n_a)
db -- Gradients of bias vector, of shape (n_a, 1)
dby -- Gradients of output bias vector, of shape (n_y, 1)
a[len(X)-1] -- the last hidden state, of shape (n_a, 1)
"""
### START CODE HERE ###
# Forward propagate through time (≈1 line)
loss, cache = rnn_forward(X, Y, a_prev, parameters)
# Backpropagate through time (≈1 line)
gradients, a = rnn_backward(X, Y, parameters, cache)
# Clip your gradients between -5 (min) and 5 (max) (≈1 line)
gradients = clip(gradients, 10)
# Update parameters (≈1 line)
parameters = update_parameters(parameters, gradients, learning_rate)
### END CODE HERE ###
return loss, gradients, a[len(X)-1]
np.random.seed(1)
vocab_size, n_a = 27, 100
a_prev = np.random.randn(n_a, 1)
Wax, Waa, Wya = np.random.randn(n_a, vocab_size), np.random.randn(n_a, n_a), np.random.randn(vocab_size, n_a)
b, by = np.random.randn(n_a, 1), np.random.randn(vocab_size, 1)
parameters = {"Wax": Wax, "Waa": Waa, "Wya": Wya, "b": b, "by": by}
X = [12,3,5,11,22,3]
Y = [4,14,11,22,25, 26]
loss, gradients, a_last = optimize(X, Y, a_prev, parameters, learning_rate = 0.01)
print("Loss =", loss)
print("gradients[\"dWaa\"][1][2] =", gradients["dWaa"][1][2])
print("np.argmax(gradients[\"dWax\"]) =", np.argmax(gradients["dWax"]))
print("gradients[\"dWya\"][1][2] =", gradients["dWya"][1][2])
print("gradients[\"db\"][4] =", gradients["db"][4])
print("gradients[\"dby\"][1] =", gradients["dby"][1])
print("a_last[4] =", a_last[4])
Loss = 126.50397572165339 gradients["dWaa"][1][2] = 0.19470931534726388 np.argmax(gradients["dWax"]) = 93 gradients["dWya"][1][2] = -0.007773876032004454 gradients["db"][4] = [-0.06809825] gradients["dby"][1] = [0.01538192] a_last[4] = [-1.]
** Expected output:**
| **Loss ** | 126.503975722 |
| **gradients["dWaa"][1][2]** | 0.194709315347 |
| **np.argmax(gradients["dWax"])** | 93 |
| **gradients["dWya"][1][2]** | -0.007773876032 |
| **gradients["db"][4]** | [-0.06809825] |
| **gradients["dby"][1]** | [ 0.01538192] |
| **a_last[4]** | [-1.] |
3.2 - Training the model¶
Given the dataset of drug names, we use each line of the dataset (one name) as one training example. Every 2000 steps of stochastic gradient descent, you will sample 7 randomly chosen names to see how the algorithm is doing. Remember to shuffle the dataset, so that stochastic gradient descent visits the examples in random order.
Exercise: Follow the instructions and implement model(). When examples[index] contains one drug name (string), to create an example (X, Y), you can use this:
index = j % len(examples)
X = [None] + [char_to_ix[ch] for ch in examples[index]]
Y = X[1:] + [char_to_ix["\n"]]
Note that we use: index= j % len(examples), where j = 1....num_iterations, to make sure that examples[index] is always a valid statement (index is smaller than len(examples)).
The first entry of X being None will be interpreted by rnn_forward() as setting $x^{\langle 0 \rangle} = \vec{0}$. Further, this ensures that Y is equal to X but shifted one step to the left, and with an additional "\n" appended to signify the end of the dinosaur name.
# GRADED FUNCTION: model
def model(data, ix_to_char, char_to_ix, num_iterations = 100001, n_a = 50, drug_names = 7, vocab_size = 27):
"""
Trains the model and generates dinosaur names.
Arguments:
data -- text corpus
ix_to_char -- dictionary that maps the index to a character
char_to_ix -- dictionary that maps a character to an index
num_iterations -- number of iterations to train the model for
n_a -- number of units of the RNN cell
drug_names -- number of drug names you want to sample at each iteration.
vocab_size -- number of unique characters found in the text, size of the vocabulary
Returns:
parameters -- learned parameters
"""
# Retrieve n_x and n_y from vocab_size
n_x, n_y = vocab_size, vocab_size
# Initialize parameters
parameters = initialize_parameters(n_a, n_x, n_y)
# Initialize loss (this is required because we want to smooth our loss, don't worry about it)
loss = get_initial_loss(vocab_size, drug_names)
# Build list of all drug names (training examples).
with open("drugnames.csv") as f:
examples = f.readlines()
examples = [x.lower().strip() for x in examples]
## some_string.partition(' ')[0]
# Shuffle list of all drug names
np.random.seed(0)
np.random.shuffle(examples)
# Initialize the hidden state of your LSTM
a_prev = np.zeros((n_a, 1))
# Optimization loop
for j in range(num_iterations):
### START CODE HERE ###
# Use the hint above to define one training example (X,Y) (≈ 2 lines)
index = j % len(examples)
X = [None] + [char_to_ix[ch] for ch in examples[index]]
Y = X[1:] + [char_to_ix["\n"]]
# Perform one optimization step: Forward-prop -> Backward-prop -> Clip -> Update parameters
# Choose a learning rate of 0.01 - experimentation shows this to produce good results.
curr_loss, gradients, a_prev = optimize(X, Y, a_prev, parameters, learning_rate = 0.005)
### END CODE HERE ###
# Use a latency trick to keep the loss smooth. It happens here to accelerate the training.
loss = smooth(loss, curr_loss)
# Every 2000 Iteration, generate "n" characters thanks to sample() to check if the model is learning properly
if j % 2000 == 0:
print('Iteration: %d, Loss: %f' % (j, loss) + '\n')
# The number of drug names to print
seed = 0
for name in range(drug_names):
# Sample indices and print them
sampled_indices = sample(parameters, char_to_ix, seed, max_length = 20)
print_sample(sampled_indices, ix_to_char)
seed += 1 # To get the same result for grading purposed, increment the seed by one.
print('\n')
return parameters
Run the following cell, you should observe your model outputting random-looking characters at the first iteration. After a few thousand iterations, your model should learn to generate reasonable-looking names.
parameters = model(data, ix_to_char, char_to_ix)
Iteration: 0, Loss: 23.080744 Nkzxwtdmfqoeyhsqwasj Kneb Kzxwtdmfqoeyhsqwasjk Neb Zxwtdmfqoeyhsqwasjkj Eb Xwtdmfqoeyhsqwasjkjv Iteration: 2000, Loss: 26.995978 Lgxttpananifvepns Inaa Ixttpamanifvepns Laa Ytsrananifvepns C Ttpananifvepns Iteration: 4000, Loss: 25.147021 Mixstodilin In Ixstodilin Mac Ytrodilhmaxerlrarele Da Trodilgnaxborobomejt Iteration: 6000, Loss: 23.882982 Onutrofilen Lleb Lvroran Ol Ytrofilen Eb Trogiine Iteration: 8000, Loss: 23.601685 Mivrpram Ine Lytrode Mecaerol Ytrode Ecagrol Utran Iteration: 10000, Loss: 23.292904 Mixtrofilin Lola Lyutoin Mecafose Yutranane Ecafprecanrevelivil Utranapiluloprasenes Iteration: 12000, Loss: 22.876617 Miwtrocimefaxaris Klec Lyutofimegax Mec Yutpan Daacose Utramaon Iteration: 14000, Loss: 22.934291 Mextrociebadulorp Inda Ivutode Mecadote Xutram Daacota Uutam Iteration: 16000, Loss: 22.592878 Mivostan Inea Lytroen Mecafore Xytramepilterlocomel Daacore Ustdiflidseros Iteration: 18000, Loss: 22.222472 Mivstramdin Inda Lytrogimed Mec Xytramclolyfort Daadose Ustaperidrestrapin Iteration: 20000, Loss: 22.345095 Mivprofilin Hlec Ivtrofilin Mec Wutram Caadote Utran Iteration: 22000, Loss: 22.072606 Mivotide Hlec Ivprodifen Mec Wutram Caadosd Utram Iteration: 24000, Loss: 21.673107 Nexstram Inda Kytrofinam Nec Vutramana Cabetrea Utramand Iteration: 26000, Loss: 21.851720 Mivoxide Hlda Iwustan Mec Vutran Caacosfab Ustaqcin Iteration: 28000, Loss: 21.687051 Onvotofidin Hlec Ivovide Ola Vprodide Caacose Ustaqcinazerip Iteration: 30000, Loss: 21.309098 Pivotral Jged Lxutrafene Pefagrol Vutramane Daadlon Ustarane Iteration: 32000, Loss: 21.434713 Nextor Hoda Jytrofime Nec Vsuram Daadose Ustarane Iteration: 34000, Loss: 21.357114 Powtroke Lola Lytrolade Phaadir Vostan Egalox Voramcin Iteration: 36000, Loss: 21.041664 Pivoxoceine Lola Lxutral Ped Votram Daadova Voseogin Iteration: 38000, Loss: 21.195486 Pivotoladin Lofa Lytroge Pedadore Votolide Daacose Vosemare Iteration: 40000, Loss: 21.144732 Piustol Lola Mytroghene Pefaine Vostan Efalovec Voral Iteration: 42000, Loss: 20.884818 Plutrol Loha Mytrol Pedadose Votran Edafose Vosenasinige Iteration: 44000, Loss: 21.003643 Pluvoschenidrespr Loic Lytroephloline Ped Votoleglimole Daacosc Voscilapatapin Iteration: 46000, Loss: 21.028819 Pluvjramane Lmed Lvovil Pegadin Votran Efagrol Voscindinidine Iteration: 48000, Loss: 20.798916 Pivotoged Mida Mytrofene Ped Votran Edacprec Vosciflimudine Iteration: 50000, Loss: 20.828868 Omytrol Kida Lyvose Ped Votram Edaklon Vosen Iteration: 52000, Loss: 20.797979 Pivovil Mlgabaton Nuvosen Pefadrol Votranasine Edagpred Vosencin Iteration: 54000, Loss: 20.736775 Plutrol Loga Mytrogelin Ped Votran Dabatref Voscilin Iteration: 56000, Loss: 20.674871 Proxypan Lola Mytro Pha Votram Ed Voscil Iteration: 58000, Loss: 20.699434 Phystogelline Miga Mytrocin Pefadose Votram Edafloma Vosenare Iteration: 60000, Loss: 20.676948 Plustide Lofa Lutrofidin Ped Votrenasile Dabatred Vosenapilol Iteration: 62000, Loss: 20.679923 Pluurofenin Kela Lyvosam Ped Votram Daacose Voram Iteration: 64000, Loss: 20.602407 Phystram Lofaacipe Mytrofensoluline Pefadrol Votramar Edakote Voralan Iteration: 66000, Loss: 20.654231 Plutrocilin Khec Luutraheprex Peg Votranale Daadrol Urodien Iteration: 68000, Loss: 20.617454 Pnovoscaine Kidaab Lyustar Ped Votran Daacor Voscelcine Iteration: 70000, Loss: 20.441042 Plustiem Kela Lutroephonavarin Pef Votran Daadose Triceflilphonibin Iteration: 72000, Loss: 20.621997 Nexotolachidrene Gelaacin Ituspan Ogabetil Votranamidine Daberol Trochene Iteration: 74000, Loss: 20.560270 Omytro Hyda Kytrofened Olabatol Votran Cabcose Urtaperidole Iteration: 76000, Loss: 20.435749 Omytrl Kida Lyutra Pha Votran Ed Tricilin Iteration: 78000, Loss: 20.591319 Olytro Hida Jutrogel Oga Votrasepole Cabdium Trocheridole Iteration: 80000, Loss: 20.579818 Omytrodide Inec Lytrocinin Peg Votran Dabetrac Trocimine Iteration: 82000, Loss: 20.316168 Omytrife Kiga Lytrolecin Olaacin Votran Dabetrid Trofletile Iteration: 84000, Loss: 20.528730 Omytro Inec Lutrokide Ogab Votran Cabbita Trofem Iteration: 86000, Loss: 20.464148 Mextrohidine Floc Hytrofilline Mbbador Votran Cabatol Trofilline Iteration: 88000, Loss: 20.220764 Omytroflin Inda Kytrogen Ogabetred Vovigil Dabesta Trocilline Iteration: 90000, Loss: 20.618712 Mextrofline Gic Hytrocilline Mec Votranalcat Cabdora Trocilline Iteration: 92000, Loss: 20.520656 Mixotranapinidine Flla Grovih Medalote Votrandin Cabetrac Trogem Iteration: 94000, Loss: 20.311747 Nexrose Gena Hytrofine Ndabetrid Vovilefa Cabetreabe Trofine Iteration: 96000, Loss: 20.396666 Mextrofimine Floc Frustan Mecaflol Votochene Cabdosa Trofeniolol Iteration: 98000, Loss: 20.528595 Omytrocindilol Glec Iutridide Olaaglor Vourandin Cabesteaclone Urogimin Iteration: 100000, Loss: 20.202982 Omytrodine Gifabatin Hytrodidin Ogabatol Vovidil Caberom Trochene
Conclusion¶
You can see that the algorithm has started to generate plausible drug names towards the end of the training. At first, it was generating random characters, but towards the end you could see drug names with definite marketing potential.
If the model generates some non-cool names, don't blame the model entirely. This experiment had used a relatively small dataset. Training a model of the english language requires a much bigger dataset, and usually needs much more computation, and could run for many hours on GPUs.
Notes¶
It may seem a little confusing what this program is doing since it is outputting results while the training phase is taking place. One of the goals of this experiment is to show how the outputs of a language model can improve with longer periods of training. So the training loop (which is happening over a total of 'n' iterations) is interrupted periodically to show how the generated drug names become more reasonable the longer the training runs.
In normal practice you would train a model, monitoring the loss to assure it is decreasing, and then at some point stop. Then you would use the model by generating some output.